Light chain
* Click for antibody sequence format fast-autocheck




GeneMedi’s LIBRA OPTMTM is a powerful optimization tool for antibody post-translational modifications (PTM) site de-risking. The LIBRA-OPTM combines sequence and structure-based in silico algorithm with antibody optimization for developing the therapeutic antibody based on post-translational modifications (PTM) site analysis, prediction and de-risking.

Post-translational modifications (PTM) refer to the modification of amino acid side chain of some proteins after biosynthesis. It is a covalent processing events, which change the properties of a protein by proteolytic cleavage and adding a modifying group, such as acetyl, phosphoryl, glycosyl and methyl, to one or more amino acids.
In recent years, the medical application of therapeutical monoclonal antibodies (mAbs) has been rapidly growing due to its targetability, low-immunogenicity, and good tolerance. It has become an important therapeutic agent for numerous diseases including cancer, autoimmune, and infectious diseases. During the therapeutical monoclonal antibody development, different physical and chemical factors influence PTM of antibody,which affect the stability, immunogenicity, and binding with target antigens.
Like all proteins, mAbs are susceptible to chemical degradation (e.g., oxidation) and enzymatic modifications (e.g., sulfation) during cell culture. Chemical and enzymatic modifications contribute to heterogeneity. For example, asparagine (Asn) deamidation could generate charge variants; tryptophan (Trp) oxidation could generate hydrophilic or hydrophobic variants. In addition, chemical modifications could affect the physical stability and biological activity of antibodies. For example, isomerization within the Fab region could reduce conformational stability, whereas deamidation within complementary-determining region (CDR) loops could reduce binding affinity.
Experimental approaches for identifying PTM liabilities are time-consuming and require high quantities of the purified protein. The sample preparation and data analysis for peptide mapping is incredibly labor intensive. At earlier stages of drug development, the number of forced degradation conditions is limited by the low availability of the purified protein. Therefore, computational tools are becoming more common during developability assessments due to the low cost, lack of sample consumption, and high speed. In the past decade, computational tools have been used to predict PTM liable sites and engineer antibodies with better chemical stability.
we focused on the common PTM of mAbs, and summarize their causes, modification sites, the influences on the mAbs’ physicochemical properties, biological activities, and stabilities, as well as the main analytical strategies, aiming to provide some references for the in-depth quality analysis of therapeutical mAbs.
Computational approaches for PTM prediction can be divided into three categories: 1. sequence-based, 2. structure-based, and 3. physics-based. Sequence-based approaches either flag individual residues prone to chemical degradation (e.g., methionine oxidation) or liable motifs (e.g., NG, NS, and NT for deamidation). Structure-based approaches predict PTM liabilities by using structural features correlated with enzymatic and chemical modifications. Common structural features include, but are not limited to, secondary structure, water coordination number (WCN), solvent-accessible surface area (SASA) and machine learning algorithms.
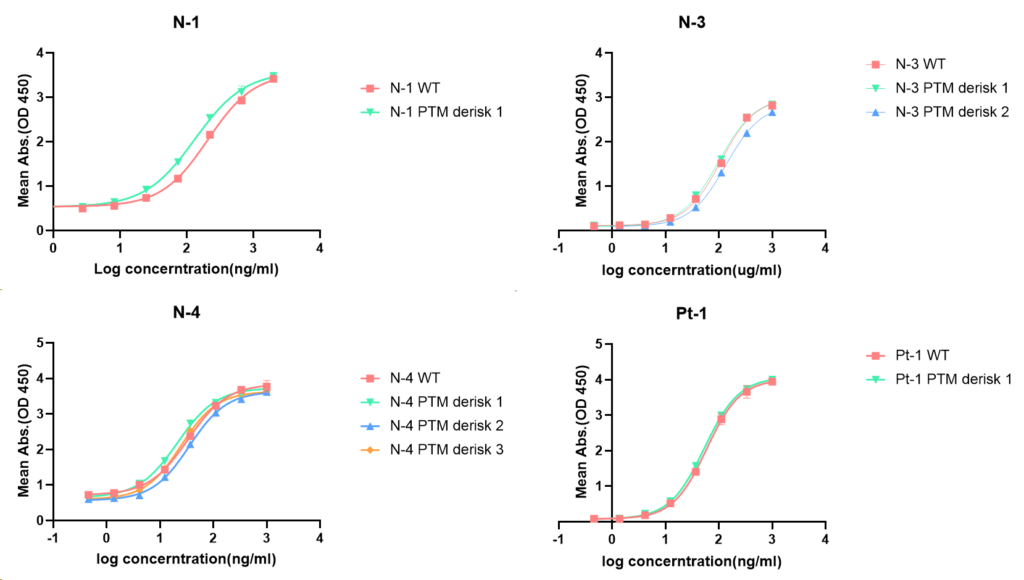
GeneMedi Biotech LIBRA-OPTM™ platform has been validated with the several antibodies of the Genemedi Biotech (Table 1.) Indirect ELISA results showed that the binding affinity of PTM de-risked antibodies were nearly similar or even higher than the native antibodies (Figure1 and Table 2). The results of this study proved that the GeneMedi Biotech LIBRA-OPTM platform is one of the powerful optimization tools for the antibody PTM site de-risking.
| Antibody Name | Antibody Cat No. | Antibody Type | Antigen Cat No. |
|---|---|---|---|
| N-1 | GMP-V-2019nCoV-NAb005 | ScFv+human IgG Fc | GMP-V-2019nCoV-N002 |
| N-3 | GMP-V-2019nCoV-NAb006 | Nanobody+human IgG Fc | GMP-V-2019nCoV-N002 |
| N-4 | GMP-V-2019nCoV-NAb007 | Human IgG1 | GMP-V-2019nCoV-N002 |
| Pt-1 | GMP-h-p-tau217-Ab01 | Mouse IgG1 | GMP-h-p-tau217-Ag01 |
Method followed
In general, LIBRA-OPTM™ provides several optimized VH/VL sequences, From this we have selected a few sequences to produce antibodies for validation. For In-direct Elisa, each well was coated with SARS-Cov-2 NP/P-tau217 for overnight at 4ºC. Followed by, it was blocked with the blocking buffer for 1 hour at 37ºC. The secondary antibody viz. HRP-conjugated anti-human IgG FC/anti-mouse IgG FC was added according to the antibody species and incubated for one hour at 37ºC. The TMB substrate was used for the color development. The color reaction was stopped using the stopping buffer and read at 450 nm using a TECAN F50 microplate reader.
Results
The table 2 showed that the most of the optimized antibodies EC50 was increased compared to the wildtype antibodies. It indicates that the PTM de-risked antibodies have a better binding affinity than the wildtype antibodies.
| Antibody | EC50(ug/ml) |
|---|---|
| N-1 WT | 207.8 |
| N-1 PTM derisk 1 | 132.7 |
| N-3 WT | 108.1 |
| N-3 PTM derisk 1 | 99.16 |
| N-3 PTM derisk 2 | 134.7 |
| N-4 WT | 33.55 |
| N-4 PTM derisk 1 | 21.29 |
| N-4 PTM derisk 2 | 35.62 |
| N-4 PTM derisk 3 | 24.94 |
| Pt-1 WT | 58.82 |
| Pt-1 PTM derisk 1 | 53.58 |


This is a demo, the antibody VH/VL sequences have been filled out. After submitting, your Email will receive a demo report.