COVID-19 related anti-2019 nCoV(SARS2 coronavirus) Antibodies for Elisa and other immunoassay anti-N protein (Nucleocapsid protein) antibody, anti-Spike protein(S protein, S1 protein, Spike RBD) antibody
Diagnostic antibodies and antigens for Companion Animal disease testing
● Rabbit
Diagnostic antibodies and antigens for Swine disease testing
Diagnostic antibodies and antigens for Avian disease testing
Diagnostic antibodies and antigens for Multiple animal disease testing
Diagnostic antibodies and antigens for Ruminant disease testing
● Deer
Diagnostic antibodies and antigens for infectious and non-infectious Equine/Horse disease testing
SOCAIL MEDIA

Content index
Introduction of GeneMedi's COVID-19 Antibodies
COVID-19, a novel pathogenic coronavirus (2019-nCoV or SARS-CoV2) induced disease, emerged in China and spread globally rapidly.
GeneMedi produces COVID-19 related anti-2019 nCoV(SARS2 coronavirus) antibodies as shown below:
Anti-2019-nCoV N protein (Nucleocapsid protein) antibody:
GMP-V-2019nCoV-NAb008
Anti-2019-nCoV NP scFv-Fc antibody:
GMP-V-2019nCoV-NAb005
Anti-2019-nCoV NP Nanobody-Fc antibody:
GMP-V-2019nCoV-NAb006
Anti-2019-nCoV NP monoclonal mouse antibody:
GMP-V-2019nCoV-NAb004
Anti-2019-nCoV-Spike protein(S protein, S1 protein, Spike RBD) antibody:
COVID-19 related antibodies (Human IgG1) were expressed in mammalian cell(human cells, HEK293).
All the antibodies of 2019 nCoV are suitable for in functional ELISA, and other immunoassays in diagnostics. The antibody can act as a capture antibody and detection antibody.
GeneMedi has submitted a preprints review about COVID-19 and 2019-nCoV.
Click here for the full-text reading:
An Insight of comparison between COVID-19 (2019-nCoV disease) and SARS in pathology and pathogenesis
In the mission of helping global scientists for the development of diagnosis and drug against 2019 nCoV(SARS2 coronavirus), GeneMedi scientists will keep moving.
2019 nCoV (SARS2 coronavirus) Antibodies for COVID-19
Cat No. | Neutralizing antibody Name | Source
(Expression Host) | Isotypes | Bioactivity validation | Order
|
|---|---|---|---|---|---|
Anti-2019-nCoV NP human monoclonal antibody |
Mammalian (human cell) |
Human IgG1 |
N protein binding,
ELISA validated as capture antibody and detection antibody
Pair recommendation with GMP-V-2019nCoV-NAb002, GMP-V-2019nCoV-NAb003, GMP-V-2019nCoV-NAb004, GMP-V-2019nCoV-NAb005, GMP-V-2019nCoV-NAb006, GMP-V-2019nCoV-NAb007, GMP-V-2019nCoV-NAb008. |
||
Anti-2019-nCoV NP human scFv-Fc antibody |
Mammalian (human cell) |
Human IgG1 |
N protein binding,
ELISA validated as capture antibody and detection antibody
Pair recommendation with GMP-V-2019nCoV-NAb001, GMP-V-2019nCoV-NAb003, GMP-V-2019nCoV-NAb004, GMP-V-2019nCoV-NAb005, GMP-V-2019nCoV-NAb006, GMP-V-2019nCoV-NAb007, GMP-V-2019nCoV-NAb008. |
||
Anti-2019-nCoV NP mouse monoclonal antibody(mAb) |
Hybridoma |
Mouse IgG |
N protein binding,
ELISA validated as capture antibody and detection antibody
Pair recommendation with GMP-V-2019nCoV-NAb001, GMP-V-2019nCoV-NAb002, GMP-V-2019nCoV-NAb004, GMP-V-2019nCoV-NAb005, GMP-V-2019nCoV-NAb006, GMP-V-2019nCoV-NAb007, GMP-V-2019nCoV-NAb008. |
||
Anti-2019-nCoV NP mouse monoclonal antibody(mAb) |
Hybridoma |
Mouse IgG |
N protein binding,
ELISA validated as capture antibody and detection antibody
Pair recommendation with GMP-V-2019nCoV-NAb001, GMP-V-2019nCoV-NAb002, GMP-V-2019nCoV-NAb003, GMP-V-2019nCoV-NAb005, GMP-V-2019nCoV-NAb006, GMP-V-2019nCoV-NAb007, GMP-V-2019nCoV-NAb008. |
||
Anti-2019-nCoV NP human scFv-Fc antibody |
Mammalian (human cell) |
Scfv |
N protein binding,
ELISA validated as capture antibody and detection antibody
Pair recommendation with GMP-V-2019nCoV-NAb001, GMP-V-2019nCoV-NAb002, GMP-V-2019nCoV-NAb003, GMP-V-2019nCoV-NAb004, GMP-V-2019nCoV-NAb006, GMP-V-2019nCoV-NAb007, GMP-V-2019nCoV-NAb008. |
||
Anti-2019-nCoV NP Nanobody-Fc antibody |
Mammalian (human cell) |
Nanobody (single-domain antibodies, sdAbs) |
N protein binding,
ELISA validated as capture antibody and detection antibody
Pair recommendation with GMP-V-2019nCoV-NAb001, GMP-V-2019nCoV-NAb002, GMP-V-2019nCoV-NAb003, GMP-V-2019nCoV-NAb004, GMP-V-2019nCoV-NAb005, GMP-V-2019nCoV-NAb007, GMP-V-2019nCoV-NAb008. |
||
Anti-2019-nCoV NP human monoclonal antibody |
Mammalian (human cell) |
human IgG |
N protein binding,
ELISA validated as capture antibody and detection antibody
Pair recommendation with GMP-V-2019nCoV-NAb001, GMP-V-2019nCoV-NAb002, GMP-V-2019nCoV-NAb003, GMP-V-2019nCoV-NAb004, GMP-V-2019nCoV-NAb005, GMP-V-2019nCoV-NAb006, GMP-V-2019nCoV-NAb008. |
||
Anti-2019-nCoV NP human monoclonal antibody |
Mammalian (human cell) |
human IgG |
N protein binding,
ELISA validated as capture antibody and detection antibody
Pair recommendation with GMP-V-2019nCoV-NAb001, GMP-V-2019nCoV-NAb002, GMP-V-2019nCoV-NAb003, GMP-V-2019nCoV-NAb004, GMP-V-2019nCoV-NAb005, GMP-V-2019nCoV-NAb006, GMP-V-2019nCoV-NAb007. |
||
Anti-2019-nCoV Spike (S1 protein) human monoclonal antibody |
Mammalian (human cell) |
Human IgG1 |
S-RBD protein binding,
ELISA validated |
||
Anti-2019-nCoV Spike (S1 protein) human monoclonal antibody |
Mammalian (human cell) |
Human IgG1 |
S-RBD protein binding,
ELISA validated,Western Blot validated |
GeneMedi's SARS-CoV-2 NP Antibody Pair And Stability Validation In Sandwich ELISA
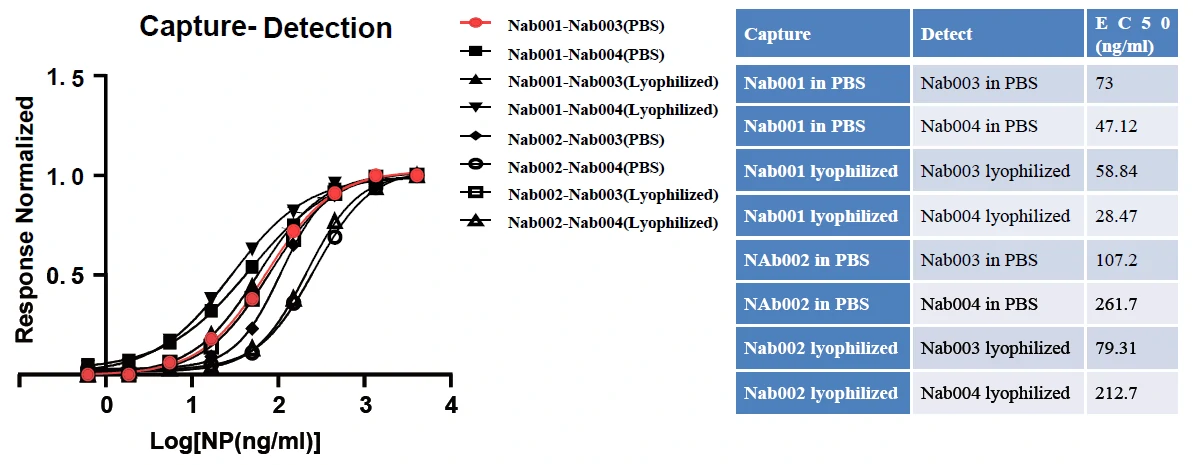
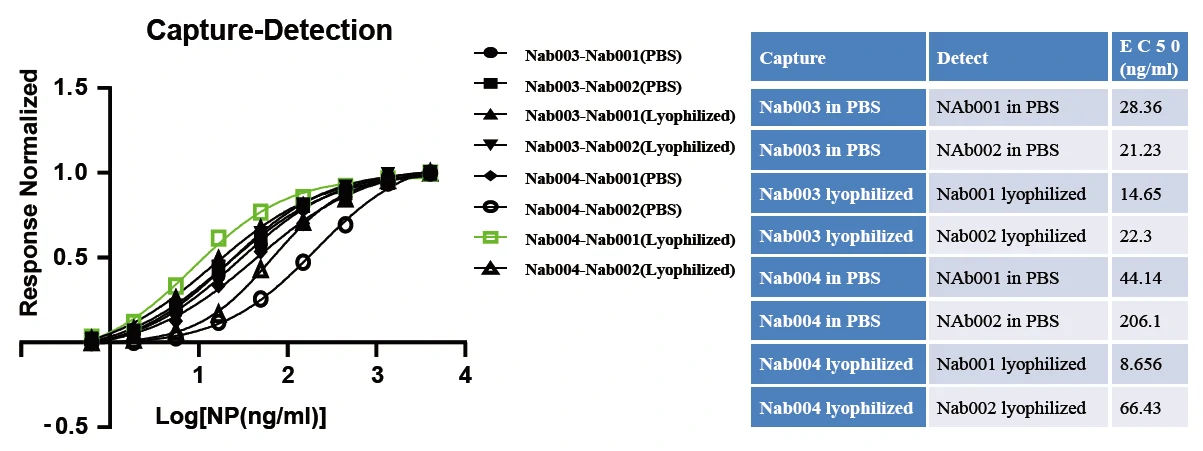
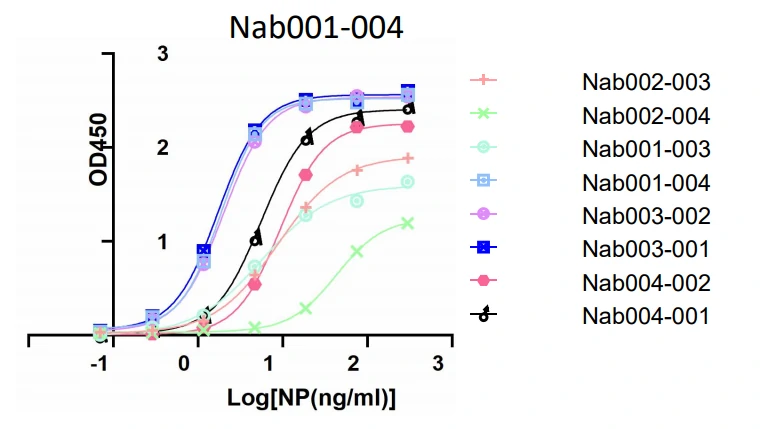
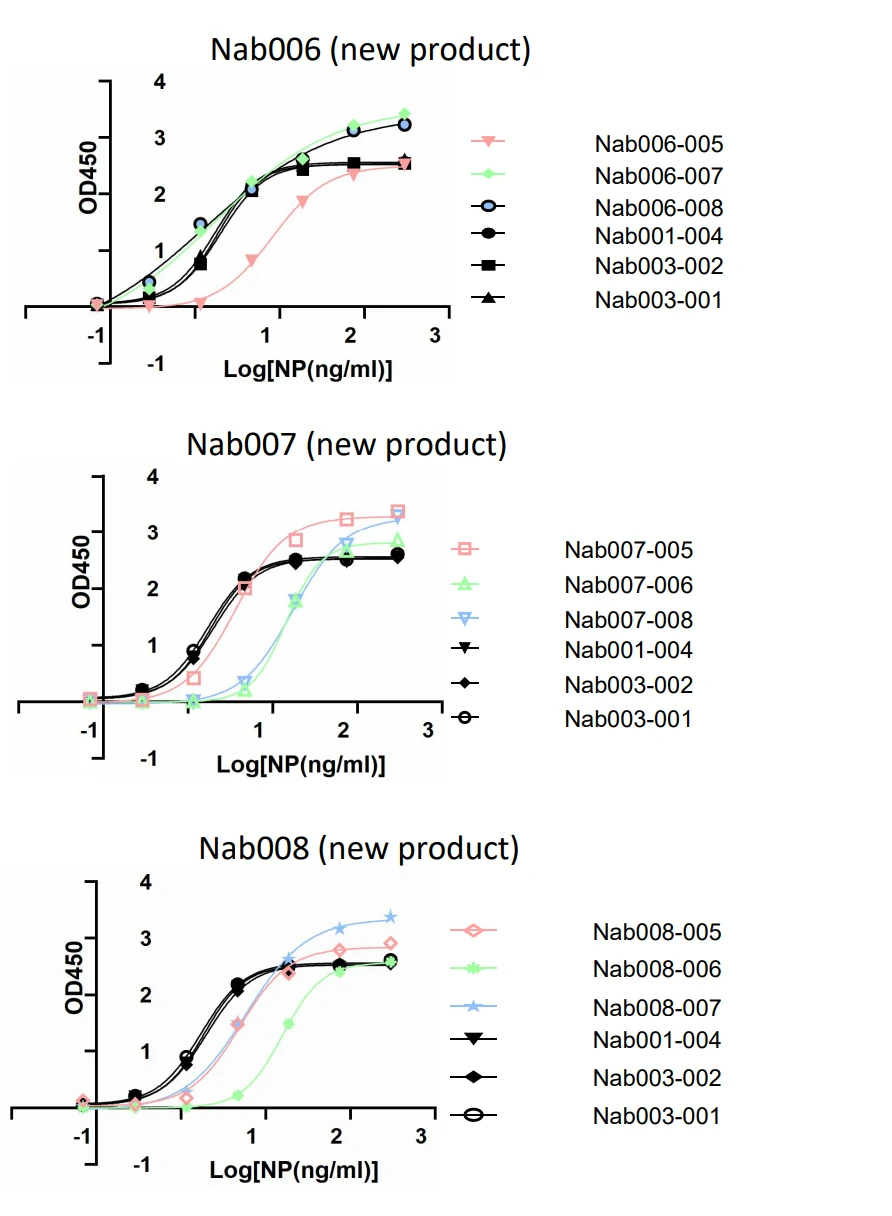
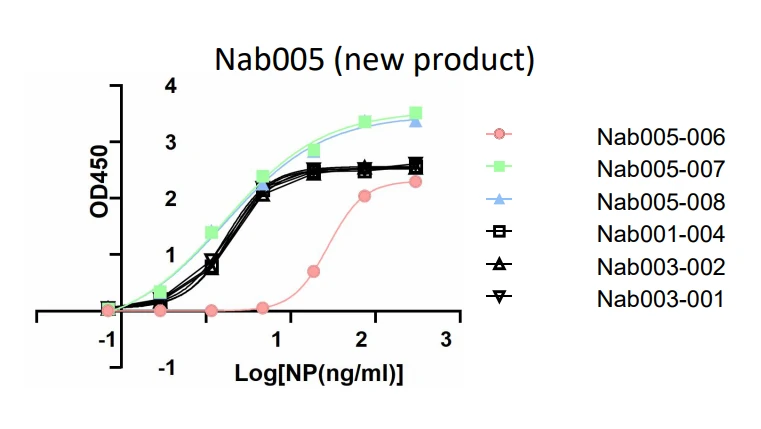
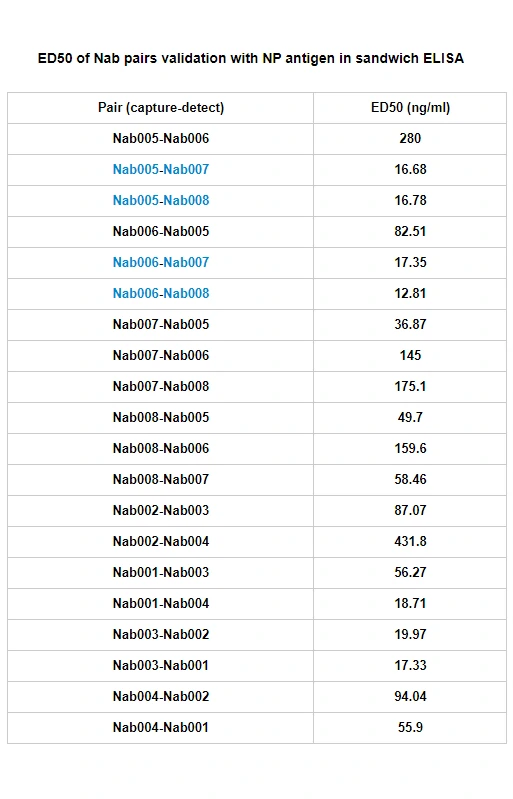
GeneMedi's SARS-CoV-2 Spike Antibody Pair And Stability Validation with Spike antigen In Sandwich ELISA


GeneMedi’s IL-6 (Interleukin 6) Antibody Pair Validation In Sandwich ELISA
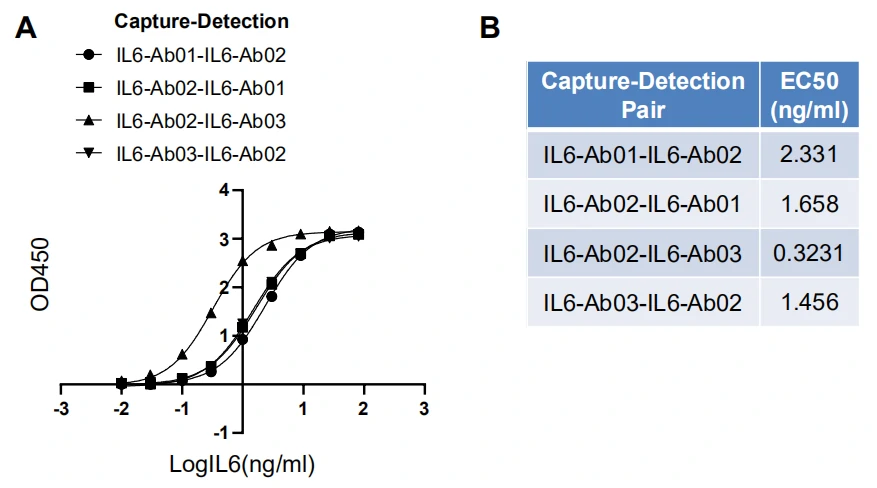
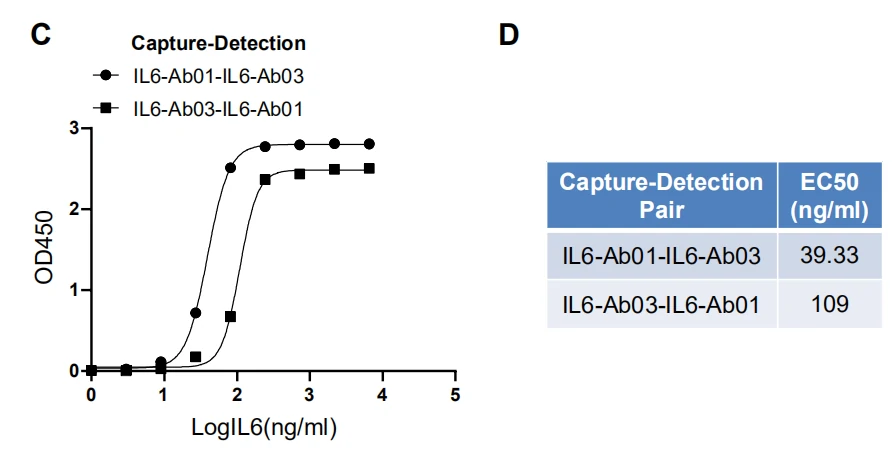
| Abbreviation | Description |
| IL6-Ab01 | GMP-h-IL6-Ab01 |
| IL6-Ab02 | GMP-h-IL6-Ab02 |
| IL6-Ab03 | GMP-h-IL6-Ab03 |
| IL6-Ag01 | GMP-h-IL6-Ag01 |
COVID-19 Products validation & publication by GeneMedi's clients
Antigens for COVID-19 Rapid Test and Antibodies Screening
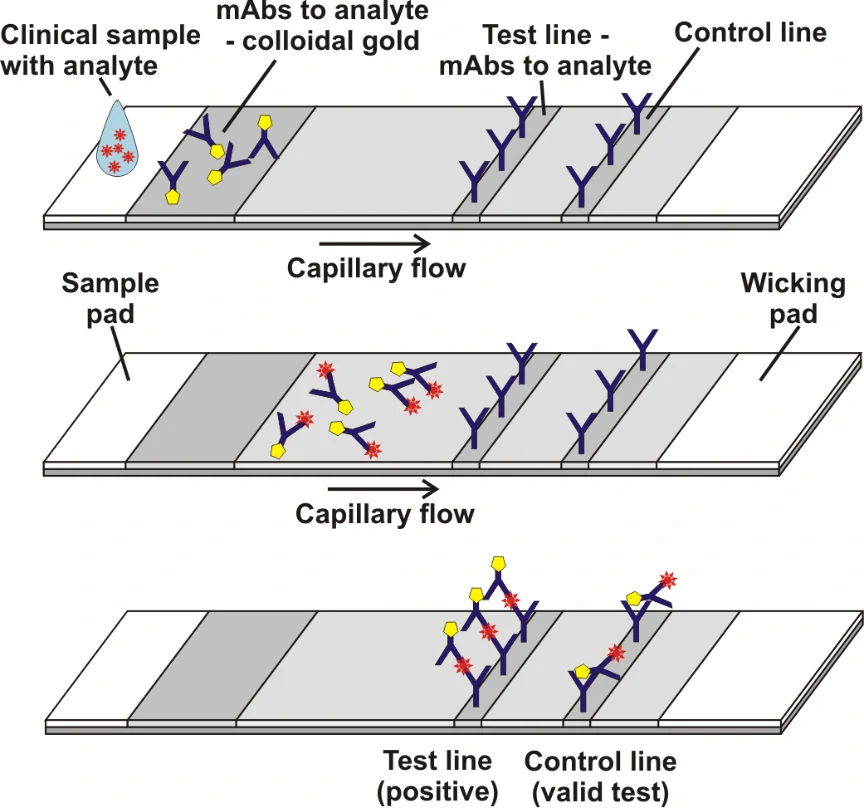
Grant et al[1]. described a half-strip Lateral flow assays (LFAs), which is a test format frequently utilized as the first step in assay development for a “full” LFA. Reverse transcription polymerase chain reaction (RT-PCR) based on oral swabs was used for confirmation of SARS-CoV-2 infection. However, high false negative rates, implementation costs and logistical problems with reagents during the global SARS-CoV-2 pandemic have hindered its universal on demand adoption. Lateral flow assays (LFAs) represent a class of diagnostic that, if sufficiently clinically sensitive, may fill many of the gaps in the current RT-PCR testing regime, especially in low- and middle-income countries.
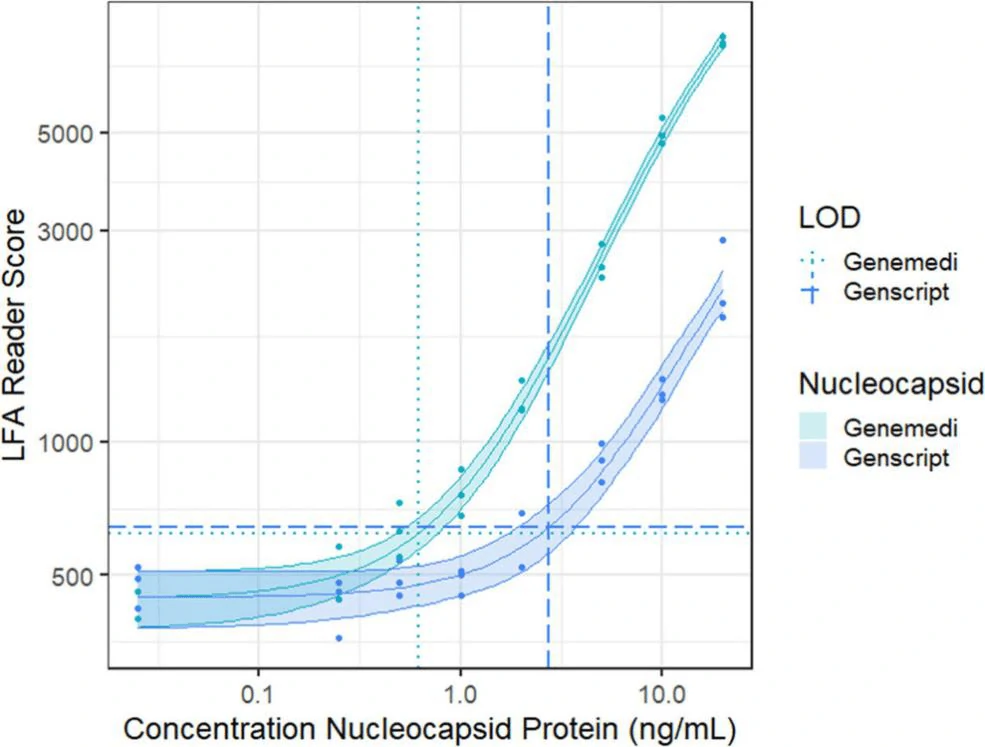
In this study, they presented a half-strip LFA for the detection of nucleocapsid protein of SARS-CoV-2. They used two commercially available SARS-CoV-2 nucleocapsid (N) proteins, from GeneMedi and Genscript to generate a dose response curve for the half-strip LFA. They found the limit of detection for the GeneMedi N protein was 0.65 ng/mL (95% CI of 0.53 to 0.77 ng/mL) and for the Genscript N protein was 3.03 ng/mL (95% CI of 0.00 to 7.44 ng/mL). The dose response curve, with 95% CI calculated in R using the drc package, is shown in Figure 2. The data indicated that the NP antigen from GeneMedi was a strong candidate for lateral flow assays (LFAs).
In another study, Cate et al [3]. described an extensive antibody screening effort that utilized high-throughput robotic antibody screening platform to screen through 673 combinations of antibody pairs that target the SARSCoV-2 nucleocapsid protein. These anti-nucleocapsid antibody pairs were tested as both capture and detection reagents with the goal of finding those pairs that have the greatest affinity for unique epitopes of the nucleocapsid protein of SARS-CoV-2 while also performing optimally in an LFA format.
Biolayer interferometry was performed on recombinant nucleocapsid proteins (NPs), for the purpose of selecting the most “native-like” analyte for LFA antibody screening. Initially, they used the estimated Rmax of five different NPs to quantify binding affinity against a random selection of 21 anti-nucleocapsid antibodies from different vendors. The metric Rmax was calculated based on theoretically saturating 100% of the bound antibody (ligand) with the analyte (NP). The NP antigen from GeneMedi was selected as the starting antigen for antibody screening because the average saturation value across 21 different anti-NP antibodies was closest to the theoretical Rmax of the antigen.
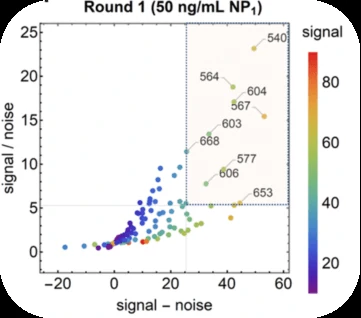
Antibody pair screening on LFAs in first-round consisted of a 11 × 11 grid of antibodies (121 unique pairs). For each pair, one antibody was striped on nitrocellulose as a test line (the “capture” antibody) and the other was coupled to latex nanoparticles using EDC/NHS chemistry (the “detector” antibody). The results of the first-round antibody pair screening on LFAs are given in Figure 5.
After the clinical based antibody pair screening, interestingly, they found the NP antigen from GeneMedi appeared to best predict antibody pair performance against clinical samples.
Antibody pairs for COVID-19 Serological Test and Immunoassay
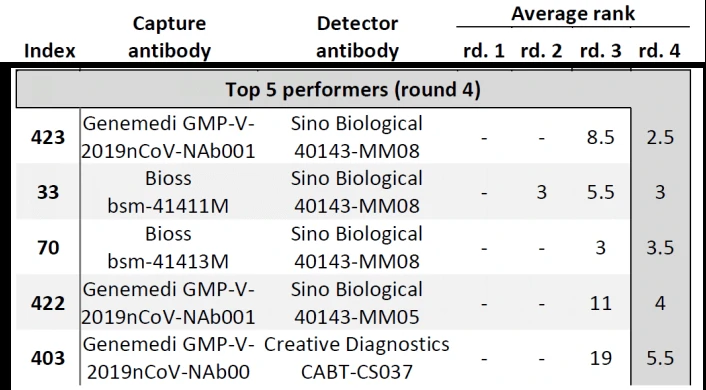
In Cate et al. ’s study[3], antibody screening results for anti-Nucleocapsid Antibodies towards the development of a SARS-CoV-2 Nucleocapsid Protein Antigen detecting Lateral Flow Assay, antibody pairs screening results in the top 5 in fourth-round from the clinical SARS-CoV-2 samples were shown in Figure 6. Among the top 5 antibody pairs, there were 3 antibody pairs from GeneMedi. This indicated that the NP antibodies from GeneMedi also behaved good performance in lateral flow assays (LFAs).
The data from these studies indicate that antigens and antibodies from GeneMedi show good performance as core ingredients of COVID-19 test kit.
[2] https://med.unr.edu/ddl/technology/lateral-flow-immunoassay
[3] Cate, David; Hsieh, Helen; Glukhova, Veronika; Bishop, Joshua D; Hermansky, H Gleda; Barrios-Lopez, Brianda; et al. (2020): Antibody Screening Results for Anti-Nucleocapsid Antibodies Towards the Development of a SARS-CoV-2 Nucleocapsid Protein Antigen Detecting Lateral Flow Assay. ChemRxiv. Preprint. https://doi.org/10.26434/chemrxiv.12709538.v1
Protocol of SARS-CoV2 (2019nCoV) NP paired antibodies sandwich ELISA
1. The Capture Antibody #1 was diluted to a concentration of 1 ~ 10 ug/ml with 0.05 M PH 9.6 carbonate buffer [1]. Then add 100 μL to each well of polystyrene plate at 4℃ overnight.
2. Aspirate the solution in the well and wash each well three times with Wash Buffer [2] (300 μL/well). Complete removal of liquid at each step is essential to good performance. Remove any remaining Wash Buffer by aspirating or decanting. Invert the plate and blot it against clean paper towels.
3. Add 5% non-fat milk and block at 37℃ for 1 h.
4. Repeat the aspiration/wash as in Step 2.
5. Add 100 μL of each serially diluted protein standard or test sample per well including blank, negative control and positive control. Ensure reagent addition is uninterrupted and completed within 15 minutes. Cover/seal the plate and incubate for 2 hours at room temperature or 37℃ for 1 hour.
6. Repeat the aspiration/wash as in Step 2.
7. Add 100 μL of Detection Antibody #2 in working concentration to each well. Cover/seal the plate and incubate for 1 hour at room temperature or 37℃ for 0.5 ~ 1 hour.
8. Repeat the aspiration/wash as in Step 2.
9. Add 200 μL of temporarily prepared TMB Substrate Solution to each well. Incubate for 20 minutes at room temperature. Protect from light.
10. Add 50 μL of 2M sulfuric acid Stop Solution to each well. If color change does not appear uniform, gently tap the plate to ensure thorough mixing.
11. Determine the optical density of each well within 20 minutes, using a microplate reader set to 450 nm. The O·D value of each well is measured after the blank control well is zeroed.
#1 Capture Antibody: Nab001 or Nab002
#2 Detection Antibody: Nab002 (if Nab001 is used as Capture Antibody) or Nab001 (if Nab002 is used as Capture Antibody)
[1] PH9.6 0.05M carbonate buffer:
Na2CO3 1.59 g
NaHCO3 2.93 g
Add distilled water to 1000ml
[2] Wash buffer (PH7.4 PBS):
KH2PO4 0.2 g
Na2HPO4·12H2O 2.9 g
NaCl 8.0 g
NaCl 0.2 g
Tween-20 0.5ml
Add distilled water to 1000ml
Protocol of SARS-CoV2 (2019nCoV) Spike-RBD antibodies standard biopanning and competitive biopanning
Standard biopanning
1. S-RBD was diluted with 0.05 M PH 9.6 carbonate buffer [1]. Then add 100 μL to each well of 96-well Maxisorp plates at 4°C overnight.
2. Aspirate the solution in the well and wash each well three times with Wash Buffer [2] (300 μL/well). Complete removal of liquid at each step is essential to good performance. Remove any remaining Wash Buffer by aspirating or decanting. Invert the plate and blot it against clean paper towels.
3. Add 1% polyvinyl alcohol (PVA) and block at room temperature for 1 hour.
4. Repeat the aspiration/wash as in Step 2.
5. Add 100μl of phage libraries (1013 pfu/ml) per well for 2 hours at room temperature.
6. Repeat the aspiration/wash eight times.
7. Add 100 μL of 100mM HCl per well for 5-min incubation to elute bound phages.
8. Transfer the eluent into a 1.5ml microfuge tube and neutralized with 1M Tris-HCl (pH 8.0).
9. Mix half the neutralized phage solution with 1ml of actively growing E. coli NEB 5-alpha F’ (OD600 = 0.8) in 2×YT media containing 10μg/ml tetracycline and incubated at 37°C for 1 hour.
10. Add 1010 pfu of M13K07 helper phages and incubated for 1 hour.
11. The infected bacteria were amplified in 50ml 2×YT medium containing 50μg/ml carbenicillin and 25μg/ml kanamycin, shaking at 200rpm and growing overnight at 37°C.
12. Phages were harvested in precipitant with PEG/NaCl solution, then resuspended in PBS buffer for the following rounds of panning.
Competitive biopanning
1. ACE2-hFc protein was diluted to a concentration of 5μg/ml with 0.05 M PH 9.6 carbonate buffer [1]. Then add 100 μL to each well of polystyrene plate at 4°C overnight.
2. Aspirate the solution in the well and wash each well three times with Wash Buffer [2]. Complete removal of liquid at each step is essential to good performance. Remove any remaining Wash Buffer by aspirating or decanting. Invert the plate and blot it against clean paper towels.
3. Add 1% polyvinyl alcohol (PVA) and block at room temperature for 1 hour.
4. Repeat the aspiration/wash as in Step 2.
5. Add the mixture of 1010 pfu antibody library and 100ng free S-RBD-His protein per well for 2-hour competitive binding at room temperature.
6. The supernatant was transferred into a 1.5ml microfuge tube containing the pre-washed Ni-NTA magnetic beads, then incubated on a shaker at room temperature for 1 hour.
7. Beads were collected using the magnetic separation rack and washed by Wash Buffer for eight times.
8. Add 100 μL of 100mM HCl per tube for 5-min incubation to elute bound phages.
9. Beads were collected using the magnetic separation rack, then transfer the eluent into a 1.5ml microfuge tube and neutralized with 1M Tris-HCl (pH 8.0).
10. Mix half the neutralized phage solution with 1ml of actively growing E. coli NEB 5-alpha F’ (OD600 = 0.8) in 2×YT media containing 10μg/ml tetracycline and incubated at 37°C for 1 hour.
11. Add 1010 pfu of M13K07 helper phages and incubated for 1 hour.
12.The bacteria were amplified on the LB plates containing 50μg/ml carbenicillin at 37°C overnight.
13. Single clones were picked up for use.
[1] PH9.6 0.05M carbonate buffer:
Na2CO3 1.59 g
NaHCO3 2.93 g
Add distilled water to 1000ml
[2] Wash buffer (PH7.4 PBS):
KH2PO4 0.2 g
Na2HPO4·12H2O 2.9 g
NaCl 8.0 g
NaCl 0.2 g
Tween-20 0.5ml
Add distilled water to 1000ml
Recombinant 2019 nCoV (SARS2 coronavirus) Antigens reagents
Cat No. | Neutralizing antibody Name | Source
(Expression Host) | Tag | Bioactivity validation | Order
|
|---|---|---|---|---|---|
Recombinant 2019-nCoV(SARS-CoV-2)
nsp10 (nsp10-CysHis,GFL protein,His Tag) |
E.coli |
N-His |
N/A |
||
Recombinant 2019-nCoV(SARS-CoV-2)
nsp16 (nsp16-OMT,2'-o-MT,His Tag) |
E.coli |
N-His |
N/A |
Potency Validated COVID-19 SARS-CoV-2 neutralizing antibody
Cat No. | Neutralizing antibody Name | Source
(Expression Host) | Isotypes | Bioactivity validation | Order
|
|---|---|---|---|---|---|
Anti-2019-nCoV Spike (Spike RBD domain) human monoclonal neutralizing antibody (IgG1) |
Mammalian (human cell) |
human IgG1 |
Validated in COVID-19 Spike protein and Spike-RBD protein binding affnity.
COVID-19 related neutralizing potency is validated by
1.2019nCoV pseudotyped virus based neutralization assay in 293T-ACE2 effector cell.
2. competitively blocking the binding of ACE-2 receptor with SARS-CoV-2 Spike protein. |
||
Anti-2019-nCoV Spike (Spike RBD domain) human monoclonal neutralizing antibody (IgM) |
Mammalian (human cell) |
human IgM |
Validated in COVID-19 Spike protein and Spike-RBD protein binding affnity.
COVID-19 related neutralizing potency is validated by
1.2019nCoV pseudotyped virus based neutralization assay in 293T-ACE2 effector cell.
2. competitively blocking the binding of ACE-2 receptor with SARS-CoV-2 Spike protein. |
||
Anti-2019-nCoV Spike (Spike RBD domain) human monoclonal neutralizing antibody (IgA) |
Mammalian (human cell) |
human IgA |
Validated in COVID-19 Spike protein and Spike-RBD protein binding affnity.
COVID-19 related neutralizing potency is validated by
1.2019nCoV pseudotyped virus based neutralization assay in 293T-ACE2 effector cell.
2. competitively blocking the binding of ACE-2 receptor with SARS-CoV-2 Spike protein. |
||
Anti-2019-nCoV Spike (Spike RBD domain) mouse monoclonal neutralizing antibody (IgG1) |
Mammalian (human cell) |
mouse IgG1 |
Validated in COVID-19 Spike protein and Spike-RBD protein binding affnity.
COVID-19 related neutralizing potency is validated by
1.2019nCoV pseudotyped virus based neutralization assay in 293T-ACE2 effector cell.
2. competitively blocking the binding of ACE-2 receptor with SARS-CoV-2 Spike protein. |
||
Anti-2019-nCoV Spike (Spike RBD domain) Cynomolgus monoclonal neutralizing antibody (IgG1) |
Mammalian (human cell) |
Cynomolgus (Non human primate, NHP) IgG1 |
Validated in COVID-19 Spike protein and Spike-RBD protein binding affnity.
COVID-19 related neutralizing potency is validated by
1.2019nCoV pseudotyped virus based neutralization assay in 293T-ACE2 effector cell.
2. competitively blocking the binding of ACE-2 receptor with SARS-CoV-2 Spike protein. |
||
Anti-2019-nCoV Spike (Spike RBD domain) human monoclonal neutralizing antibody (IgG1) |
Mammalian (human cell) |
human IgG1 |
Validated in COVID-19 Spike protein and Spike-RBD protein binding affnity.COVID-19 related neutralizing potency is validated by 1.2019nCoV pseudotyped virus based neutralization assay in 293T-ACE2 effector cell. 2. competitively blocking the binding of ACE-2 receptor with SARS-CoV-2 Spike protein. |
||
Anti-2019-nCoV Spike (Spike RBD domain) human monoclonal neutralizing antibody (IgG1) |
Mammalian (human cell) |
human IgG1 |
Validated in COVID-19 Spike protein and Spike-RBD protein binding affnity.
COVID-19 related neutralizing potency is validated by
1.2019nCoV pseudotyped virus based neutralization assay in 293T-ACE2 effector cell.
2. competitively blocking the binding of ACE-2 receptor with SARS-CoV-2 Spike protein. |
||
Anti-2019-nCoV Spike (Spike RBD domain) human monoclonal neutralizing antibody (IgG1) |
Mammalian (human cell) |
human IgG1 |
Validated in COVID-19 Spike protein and Spike-RBD protein binding affnity.COVID-19 related neutralizing potency is validated by 1.2019nCoV pseudotyped virus based neutralization assay in 293T-ACE2 effector cell. 2. competitively blocking the binding of ACE-2 receptor with SARS-CoV-2 Spike protein. |
SARS-CoV-2 neutralizing antibody validated post download
–Nab discovery and vaccines evaluation through SARS-CoV-2 wildtype/mutant variants pseudovirus based neutralizing assay(PBNA) and Spike-ACE2 competition binding assay
GeneMedi-SARS-CoV-2 WT and Spike Mutation Variants Pseudovirus (PSV) Based Cell Entry

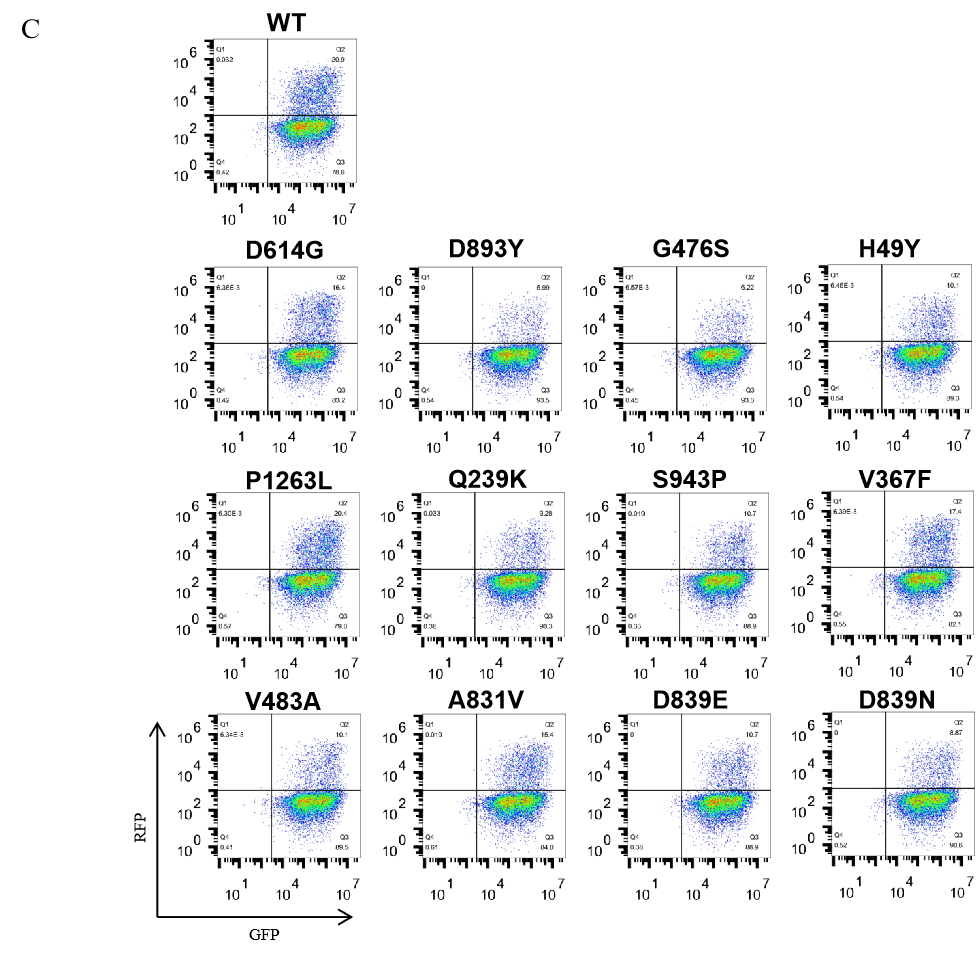
GeneMedi's anti-2019-nCoV Spike Neutralizing antibodies (Nabs) and Spike RBD protein binding validation
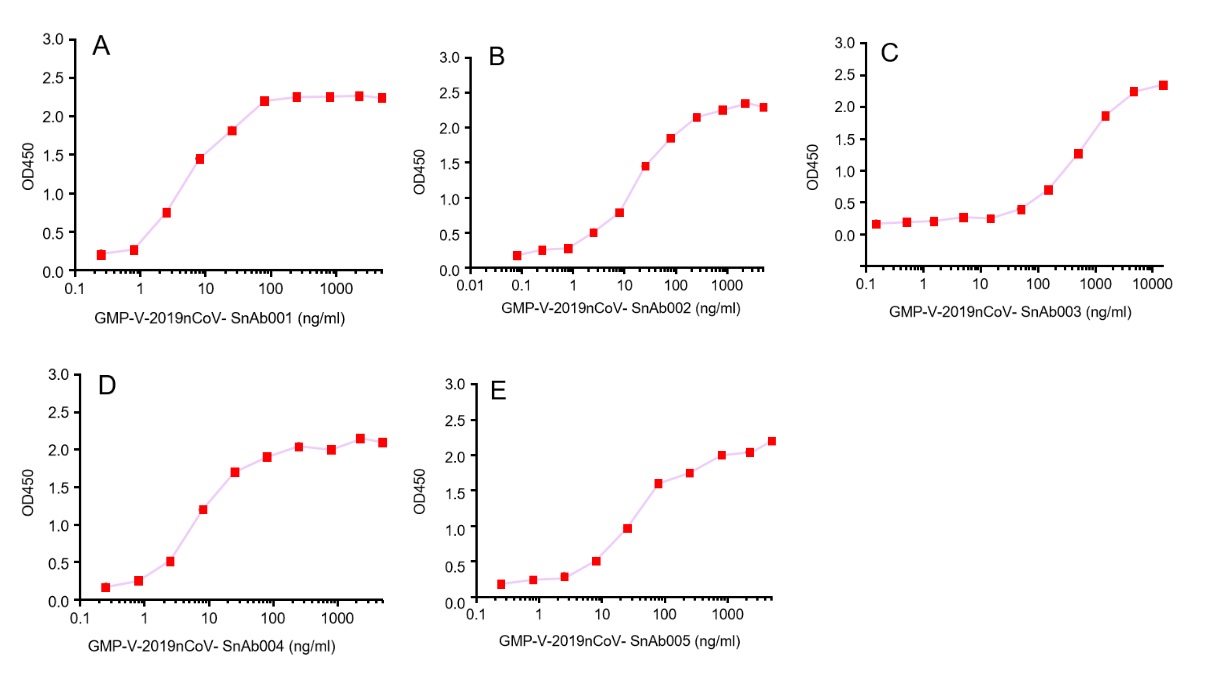
| Cat No. | Product | EC50 (ng/ml) |
| GMP-V-2019nCoV-SnAb001 | Anti-2019-nCoV Spike (Spike RBD domain) human monoclonal neutralizing antibody (IgG1) | 5 |
| GMP-V-2019nCoV-SnAb002 | Anti-2019-nCoV Spike (Spike RBD domain) human monoclonal neutralizing antibody (IgM) | 18 |
| GMP-V-2019nCoV-SnAb003 | Anti-2019-nCoV Spike (Spike RBD domain) human monoclonal neutralizing antibody (IgA) | 410 |
| GMP-V-2019nCoV-SnAb004 | Anti-2019-nCoV Spike (Spike RBD domain) mouse monoclonal neutralizing antibody (IgG1) | 6.8 |
| GMP-V-2019nCoV-SnAb005 | Anti-2019-nCoV Spike (Spike RBD domain) Cynomolgus monoclonal neutralizing antibody (IgG1) | 28 |
A.GMP-V-2019nCoV-SnAb001:Anti-2019-nCoV Spike (Spike RBD domain) human monoclonal neutralizing antibody (IgG1)
B.GMP-V-2019nCoV-SnAb002:Anti-2019-nCoV Spike (Spike RBD domain) human monoclonal neutralizing antibody (IgM)
C.GMP-V-2019nCoV-SnAb003:Anti-2019-nCoV Spike (Spike RBD domain) human monoclonal neutralizing antibody (IgA)
D.GMP-V-2019nCoV-SnAb004:Anti-2019-nCoV Spike (Spike RBD domain) mouse monoclonal neutralizing antibody (IgG1)
E.GMP-V-2019nCoV-SnAb005:Anti-2019-nCoV Spike (Spike RBD domain) Cynomolgus monoclonal neutralizing antibody (IgG1)
GeneMedi's anti-2019-nCoV Spike Neutralizing antibodies (Nabs) competitive binding assay validation
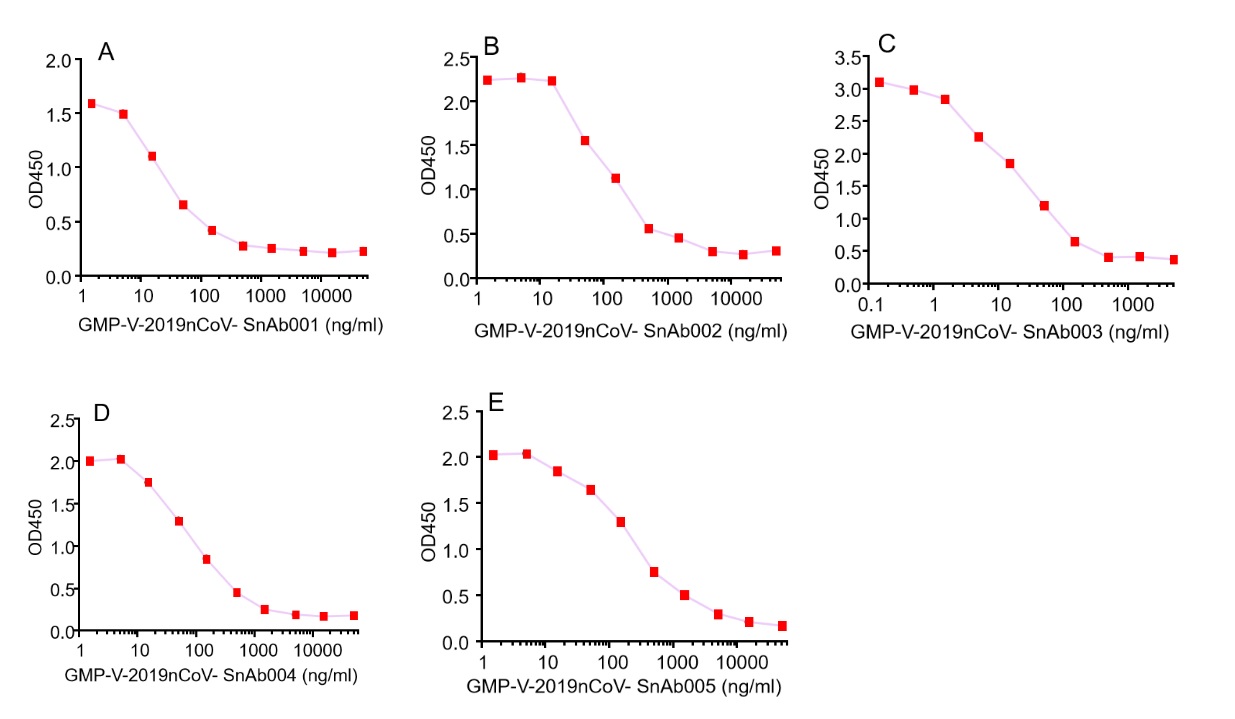
| Cat No. | Product | IC50 (ng/ml) |
| GMP-V-2019nCoV-SnAb001 | Anti-2019-nCoV Spike (Spike RBD domain) human monoclonal neutralizing antibody (IgG1) | 26.3 |
| GMP-V-2019nCoV-SnAb002 | Anti-2019-nCoV Spike (Spike RBD domain) human monoclonal neutralizing antibody (IgM) | 84.2 |
| GMP-V-2019nCoV-SnAb003 | Anti-2019-nCoV Spike (Spike RBD domain) human monoclonal neutralizing antibody (IgA) | 20.5 |
| GMP-V-2019nCoV-SnAb004 | Anti-2019-nCoV Spike (Spike RBD domain) mouse monoclonal neutralizing antibody (IgG1) | 81.9 |
| GMP-V-2019nCoV-SnAb005 | Anti-2019-nCoV Spike (Spike RBD domain) Cynomolgus monoclonal neutralizing antibody (IgG1) | 243 |
GeneMedi-SARS-CoV-2 WT and Spike Mutation Variants Pseudovirus (PSV) Based Neutralizing Assay with GeneMedi's anti-2019-nCoV Spike Neutralizing antibodies (Nabs)


About SARS-CoV-2 (2019nCoV, novel Coronavirus) Spike protein, S1 Protein, Spike S1-NTD, Spike S1-CTD, Spike-RBD and Spike trimer protein.
1. SARS-CoV-2 (2019nCoV) Spike protein: SARS-CoV-2, a newly emerged pathogen spreading worldwide. The transmembrane spike (S) glycoprotein of SARS-CoV-2 that forms homotrimers protruding from the viral surface is known to mediate coronavirus entry into host cells. It has been reported that spike protein can bind with high affinity to human ACE2 and uses it as an entry receptor to invade target cells.
2. SARS-CoV-2 (2019nCoV) spike S1 protein: The spike protein is a large type I transmembrane glycoprotein comprises two functional subunits, S1 and S2. S1 subunit of spike protein is responsible for binding to the host cell receptor. S2 subunit is responsible for fusion of the viral and cellular membranes.
3. SARS-CoV-2 (2019nCoV) spike S1-NTD and S1-CTD: For most coronaviruses, the N-terminal domain (NTD) of the S1 subunit attaches to cellular carbohydrates and the C-terminal domain of S1 (S1-CTD) binds to a cellular protein receptor. Carbohydrate binding by the S1 N-terminal domain is thought to keep the virus in close proximity to the host cell surface, whereas engagement of specific protein receptors by the S1-CTD is thought to initiate a series of conformational changes in the spike that ultimately result in membrane fusion and delivery of the viral genome to the cytosol.
4. SARS-CoV-2 (2019nCoV) Spike RBD: S1 subunit of spike protein contains a receptor binding domain (RBD), which is responsible for recognizing the cell surface receptor.
5. SARS-CoV-2 (2019nCoV) Spike trimer protein: The transmembrane spike (S) glycoprotein of SARS-CoV-2 usually forms homotrimers protruding from the viral surface. S trimers are extensively decorated with N-linked glycans that are important for proper folding and for modulating accessibility to host proteases and neutralizing Abs. It has been found that S glycoprotein trimers in highly pathogenic human coronaviruses appear to exist in partially opened states, while they remain largely closed in human coronaviruses associated with common colds.
About SARS-CoV-2 (2019nCoV, novel Coronavirus) Envelop protein (Coronavirus E protein)
1. SARS-CoV-2 (2019nCoV) E protein: E protein is the smallest major structural proteins. It has a N-terminal ectodomain and a C-terminal endodomain with ion channel activity. During the replication cycle, E protein is abundantly expressed inside the infected cell, but only a small portion is incorporated into the virus envelope. The majority of the protein participates in viral assembly and budding. E protein is important in virus production and maturation. Recombinant CoVs without E have been shown to exhibit significantly reduced viral titres, crippled viral maturation, or yield incompetent progeny.
About SARS-CoV-2 (2019nCoV, novel Coronavirus) membrane protein (Coronavirus M protein)
1. SARS-CoV-2 (2019nCoV) M protein: Coronavirus M protein is believed to define the shape of the viral envelope,which contains three transmembrane domains. It has a small N-terminal glycosylated ectodomain and a much larger C-terminal endodomain that extends 6–8 nm into the viral particle. M protein usualy exists as a dimer, and may adopt two different conformations allowing it to promote membrane curvature as well as bind to the nucleocapsid.
About SARS-CoV-2 (2019nCoV, novel Coronavirus) Nucleocapsid protein (Coronavirus N protein)
1. SARS-CoV-2 (2019nCoV) N protein: Coronavirus N protein is required for coronavirus RNA synthesis, and has RNA chaperone activity that may be involved in template switch. Nucleocapsid protein is a most abundant protein of coronavirus. N protein packages the positive strand viral genome RNA into a helical ribonucleocapsid (RNP) and plays a fundamental role during virion assembly through its interactions with the viral genome and membrane protein M. Plays an important role in enhancing the efficiency of subgenomic viral RNA transcription as well as viral replication. Because of the conservation of N protein sequence and its strong immunogenicity, the N protein of coronavirus is chosen as a diagnostic tool.
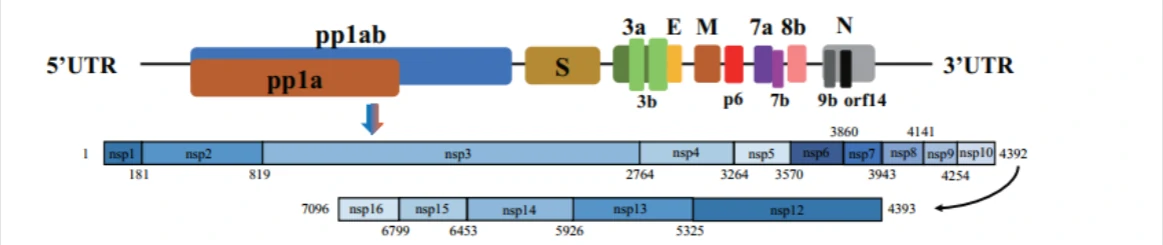
About SARS-CoV-2 (2019nCoV, novel Coronavirus)Non-structure protein (Nsp1-Nsp16)
2019nCoV contains 16 Non-structure protein (Nsp1-Nsp16) that may be drugable targets for antiviral compounds discovery against COVID-191.
| Non-structure proteins | Starting position (aa) | Ending position (aa) | Length (aa) |
| nsp1 | 1 | 180 | 180 |
| (leader protein) | |||
| nsp2 | 181 | 818 | 638 |
| nsp3 | 819 | 2763 | 1945 |
| (Papain-Like proteinase, PLpro) | |||
| nsp4 | 2764 | 3263 | 500 |
| nsp5 | 3264 | 3569 | 306 |
| (Mpro, Main proteinase, 3C-like proteinase) | |||
| nsp6 | 3570 | 3859 | 290 |
| nsp7 | 3860 | 3942 | 83 |
| nsp8 | 3943 | 4140 | 198 |
| nsp9 | 4141 | 4253 | 113 |
| nsp10 | 4254 | 4392 | 139 |
| (growth-factor-like protein) | |||
| nsp12 | 4393 | 5324 | 932 |
| (RdRP,NA-dependent RNA polymerase) | |||
| nsp13 | 5325 | 5925 | 601 |
| (RNA 5′-triphosphatase) | |||
| nsp14 | 5926 | 6452 | 527 |
| (3′-to-5′ exonuclease) | |||
| nsp15 | 6453 | 6798 | 346 |
| (endoRNAse) | |||
| nsp16 | 6799 | 7096 | 298 |
| (2’O-MTase, 2′-O-ribose methyltransferase) |
1. Nsp3: Nsp3 (200 kDa) is the largest protein encoded by the coronavirus (CoV) genome. Nsp3 is an essential component of the replication and transcription complex. It comprises various domains, the organization of which differs between CoV genera, due to duplication or absence of some domains. However, the N-terminal region of the Nsp3 is highly conserved among CoV, containing a ubiquitin-like (Ubl) globular fold followed by a flexible, extended acidic-domain (AC domain) rich in glutamic acid (38%). Next to the AC domain is a catalytically active ADP-ribose-1″-phosphatase (ADRP, app-1″-pase) domain (also called macro domain or X domain) thought to play a role during synthesis of viral subgenomic RNAs. SARS Unique Domain (SUD), a domain not yet identified in other coronaviruses from alphacoronavirus and betacoronavirus, follows next. The SUD domain binds oligonucleotides known to form G-quadruplexes. Downstream of the SUD domain is a second Ubl domain and the catalytically active PLpro domain that proteolytically processes the Nsp1/2, Nsp2/3 and Nsp3/4 cleavage sites. Downstream of PLpro are found a nucleic acid-binding domain (NAB) with a nucleic acid chaperon function, which is conserved in betacoronavirus and gammacoronavirus, and one uncharacterized domain termed the marker domain (G2M). Following the G2M are two predicted double-pass transmembrane domains (TM1–2 and TM3–4), a putative metal binding region (ZN) and the Y domain of unknown function (subdomains Y1–3).
2. Nsp5: Nsp5 protease (3CLpro; Mpro) mediates processing at 11 distinct cleavage sites, including its own autoproteolysis, and is essential for virus replication. Nsp5 exhibits a conserved three-domain structure and catalytic residues.
3. Nsp10: Nsp10 (18 kDa) is well conserved among coronaviruses and encoded by ORF1a. It’s thought to serve as an important multifunctional cofactor in replication. Nsp10 was shown to interact with itself, as well as with Nsp1, Nsp7, Nsp14, and Nsp16. The important role of Nsp10 is responsible for RNA synthesis. It was shown that a murine hepatitis virus (MHV) temperature-sensitive mutant carrying a non-synonymous mutation in the Nsp10 coding sequence had a defect in minus-strand RNA synthesis at non-permissive temperatures.
4. Nsp12: Nsp12 (102 kDa) is a multidomain RNA polymerase, which is the most conserved protein in coronaviruses. Nsp12 contains an RNA-dependent RNA polymerase (RdRp) domain in its C-terminal, which is essential for the viral replication and transcription.
5. Nsp16: Nsp16 is an SAM-dependent nucleoside-2’O-methyl-transferase (2’O-MTase). The mRNA cap for coronaviruses is completed by Nsp16, which ensures formation of a protective cap-1 structure that prevent recognition by either MDA5 or IFIT proteins. Finally, the NSP16/NSP10 complex finishes coronavirus capping process permitting viral infection with reduced host recognition.
About COVID-19 pandemic, Coronavirus (Coronavirus) and genome of SARS-CoV-2 (2019nCoV)
COVID-19 pandemic is caused by 2019nCoV (SARS-CoV-2, a novel coronavirus) infection.The 2019-nCoV genome was annotated to possess 14 ORFs encoding 27 proteins1.
| Gene name of 2019nCoV (SARS-CoV-2, a novel coronavirus) | Coding region(nt) | Protein length(aa) |
| orf1a | 266-13483 | 4405 |
| orf1ab | 266-13468, 13468-21555 | 7096 |
| S | 21563-25384 | 1273 |
| 3a | 25393-26220 | 275 |
| 3b | 25814-25882 | 22 |
| 26183-26281 | 32 | |
| E (envelope protei) | 26245-26472 | 75 |
| M (matrix protein) | 26523-27191 | 222 |
| p6 | 27202-27387 | 61 |
| 7a | 27394-27759 | 121 |
| 7b | 27756-27887 | 43 |
| 8b | 27894-28259 | 121 |
| 9b | 28284-28577 | 97 |
| N(nucleocapsid) | 28274-29533 | 419 |
| orf14 | 28734-28955 | 73 |
Collection of COVID-19 landscape knowledge base
1.Wu, A. et al. Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China. Cell Host Microbe, doi:10.1016/j.chom.2020.02.001 (2020).




