RT-PCR (Real time -PCR) in Coronavirus Disease 2019 (COVID-19) test
Diagnostic antibodies and antigens for Companion Animal disease testing
● Rabbit
Diagnostic antibodies and antigens for Swine disease testing
Diagnostic antibodies and antigens for Avian disease testing
Diagnostic antibodies and antigens for Multiple animal disease testing
Diagnostic antibodies and antigens for Ruminant disease testing
● Deer
Diagnostic antibodies and antigens for infectious and non-infectious Equine/Horse disease testing
SOCAIL MEDIA

Author:
Huajun Bai1 Xiaolong Cai1, 2* Xiaoyan Zhang1
1. R&D Center, GeneMedi Co.Ltd., Shanghai, P.R. China (www.genemedi.com)
2. Hanbio Research Center, Hanbio Tech Co. Ltd., Shanghai, P.R. China (www.hanbio.net)
Abstract:
The outbreak of COVID-19, caused by 2019 novel coronavirus (2019-nCoV), has been a global public health threat and caught the worldwide concern. Scientists throughout the world are sparing all efforts to explore strategies for the determination of the 2019-nCoV virus and diagnosis of COVID-19 rapidly. Several assays are developed for COVID-19 test , including RT-PCR, coronavirus antigens-based immunoassays, and CRISPR-based strategies (Cas13a or Cas12a), etc. Different assays have their advantages and drawbacks, and people should choose the most suitable assay according to their demands. Here, we make a brief introduction about these assays and give a simple overview of them, hoping to help doctors and researchers to select the most suitable assay for the Coronavirus Disease 2019 test (COVID-19 test) .
RT-PCR (Real time -PCR) in Coronavirus Disease 2019 (COVID-19) test
1) Principles for diagnostics
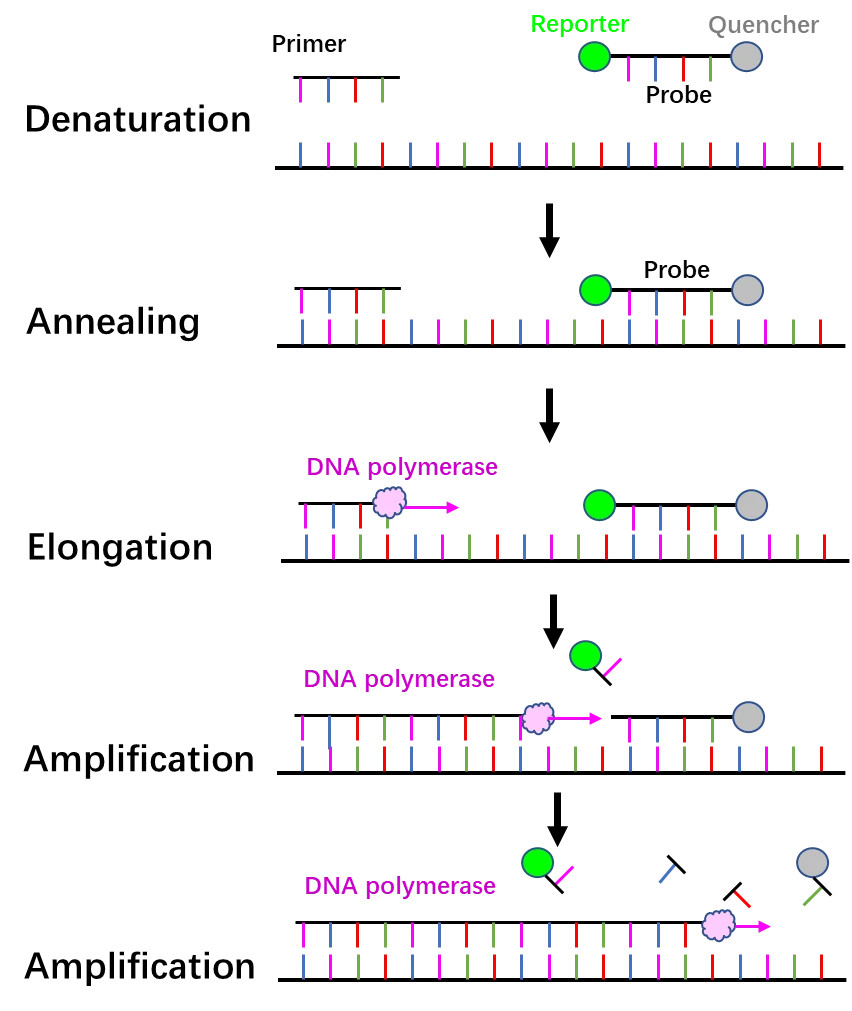
2) Advantages and disadvantages
Advantages:
① High sensitivity.
② Easy to be operated on a large scale.
Disadvantages:
① Professional technician and special apparatus are required to perform RT-PCR experiments and analyze the data, which costs so much.
② The standard positive control affects the experimental accuracy, and a false negative result may occur due to improper handling.
3) Optional Targets, primers, and probes from different departments
Different departments have their own systems for the determination of COVID-19 by targeting various open reading frames of SARS-CoV-2, which are shown as follows. Primers and probes of Orf1ab and N (nucleocapsid gene) are commonly used for COVID-19 test in China. Three pairs of primers and probes of N (nucleocapsid gene, N1, N2, N3) are applied in the US CDC for COVID-19. And primers and probes of E (envelope gene) are utilized for COVID-19 test in Europe.
| Organisation | Target | Forward primer (5’-3’) | Reverse primer (5’-3’) | Probe (5’-3’) |
University of Hong Kong, Beijing Center for Disease Prevention and Control, Capital Medical University Beijing Research Center for Preventive Medicine | orf1b | TGGGGYTTTACRGGTAACCT (Forward; Y=C/T, R=A/G) | AACRCGCTTAACAAAGCACTC (Reverse; R=A/G) | TAGTTGTGATGCWATCATGACTAG (Probe in 5’-FAM/ZEN/3’-IBFQ format; W=A/T) |
| N | TAATCAGACAAGGAACTGATTA | CGAAGGTGTGACTTCCATG | FAM-GCAAATTGTGCAATTTGCGG-IBFQ | |
Charité Virology, Berlin, Germany; Olfert Landt, Tib-Molbiol, Berlin, Germany; Erasmus MC, Rotterdam, The Netherlands; Public Health England, London | RdRP | GTGARATGGTCATGT GTGGCGG | CARATGTTAAASACA CTATTAGCATA | FAM-CAGGTGGA ACCTCATCAGGA GATGCBBQ |
| E gene | ACAGGTACGTTAATA GTTAATAGCGT | ATATTGCAGCAGTAC GCACACA | FAM-ACACTAGC CATCCTTACTGC GCTTCGBBQ | |
| N | CACATTGGCACCCGC AATC | GAGGAACGAGAAGA GGCTTG | FAM-ACTTCCTCA AGGAACAACATT GCCABBQ | |
The US Centers for Disease Control and Prevention (CDC), Integrated DNA Technologies | N1 | GACCCCAAAATCAGC GAAAT | TCTGGTTACTGCCAG TTG AATCTG | FAM-ACCCCGCA T TACGTTTGGTGG ACC-BHQ1 |
| N2 | TTACAAACATTGGCCGCAAA | GCGCGACATTCCGAAGAA | FAM-ACAATTTGCCCCCAGCGCTTCAG-BHQ1 | |
| N3 | GGGAGCCTTGAATACACCAAAA | TGTAGCACGATTGCAGCATTG | FAM-AYCACATTGGCACCCGCAATCCTG-BHQ1 | |
| RNAse P | AGATTTGGACCTGCGAGCG | GAGCGGCTGTCTCCACAAGT | FAM–TTCTGACCTGAAGGCTCTGCGCG–BHQ-1 |
Table 1. Optional Targets, primers, and probes from different departments
Reference
1.L.E.a.V.D.M. Gralinski, Return of the Coronavirus: 2019-nCoV. , Viruses, 2020. 12(2). (2020).
2.J.L. V. M. Corman, M. Witzenrath, Coronaviruses as the cause of respiratory infections, Internist (Berl) 60, 1136-1145 (2019).
3.Y. Yang, Q. Lu, M. Liu, Y. Wang, A. Zhang, N. Jalali, N. Dean, I. Longini, M.E. Halloran, B. Xu, X. Zhang, L. Wang, W. Liu, L. Fang, Epidemiological and clinical features of the 2019 novel coronavirus outbreak in China, medRxiv, (2020).
4.J. Li, S. Li, Y. Cai, Q. Liu, X. Li, Z. Zeng, Y. Chu, F. Zhu, F. Zeng, Epidemiological and Clinical Characteristics of 17 Hospitalized Patients with 2019 Novel Coronavirus Infections Outside Wuhan, China, medRxiv, (2020).
5.C. Huang, Y. Wang, X. Li, L. Ren, J. Zhao, Y. Hu, L. Zhang, G. Fan, J. Xu, X. Gu, Z. Cheng, T. Yu, J. Xia, Y. Wei, W. Wu, X. Xie, W. Yin, H. Li, M. Liu, Y. Xiao, H. Gao, L. Guo, J. Xie, G. Wang, R. Jiang, Z. Gao, Q. Jin, J. Wang, B. Cao, Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China, The Lancet, 395 (2020) 497-506.
6.W.-j. Guan, Z.-y. Ni, Y. Hu, W.-h. Liang, C.-q. Ou, J.-x. He, L. Liu, H. Shan, C.-l. Lei, D.S.C. Hui, B. Du, L.-j. Li, G. Zeng, K.-Y. Yuen, R.-c. Chen, C.-l. Tang, T. Wang, P.-y. Chen, J. Xiang, S.-y. Li, J.-l. Wang, Z.-j. Liang, Y.-x. Peng, L. Wei, Y. Liu, Y.-h. Hu, P. Peng, J.-m. Wang, J.-y. Liu, Z. Chen, G. Li, Z.-j. Zheng, S.-q. Qiu, J. Luo, C.-j. Ye, S.-y. Zhu, N.-s. Zhong, Clinical characteristics of 2019 novel coronavirus infection in China, medRxiv, (2020).
7.N. Chen, M. Zhou, X. Dong, J. Qu, F. Gong, Y. Han, Y. Qiu, J. Wang, Y. Liu, Y. Wei, J.a. Xia, T. Yu, X. Zhang, L. Zhang, Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study, The Lancet, 395 (2020) 507-513.
8.A. Wu, Y. Peng, B. Huang, X. Ding, X. Wang, P. Niu, J. Meng, Z. Zhu, Z. Zhang, J. Wang, J. Sheng, L. Quan, Z. Xia, W. Tan, G. Cheng, T. Jiang, Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China, Cell Host Microbe, (2020).
9.R.C. A. R. Fehr, S. Perlman,, Middle East Respiratory Syndrome:Emergence of a Pathogenic Human Coronavirus, Annu Rev Med 68, 387-399 (2017).
10.X.Y. Ge, Li, J.L., Yang, X.L., Chmura, A.A., Zhu, G.,Epstein, J.H., Mazet, J.K., Hu, B., Zhang, W., Peng,C., et al. , Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor., Nature 503, 535–538 (2013).
11.M. Hoffmann, H. Kleine-Weber, N. Krüger, M. Müller, C. Drosten, S. Pöhlmann, The novel coronavirus 2019 (2019-nCoV) uses the SARS-coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells, bioRxiv, (2020).
12.C. Fan, K. Li, Y. Ding, W.L. Lu, J. Wang, ACE2 Expression in Kidney and Testis May Cause Kidney and Testis Damage After 2019-nCoV Infection, medRxiv, (2020).
13.R. Channappanavar, C. Fett, M. Mack, P.P. Ten Eyck, D.K. Meyerholz, S. Perlman, Sex-Based Differences in Susceptibility to Severe Acute Respiratory Syndrome Coronavirus Infection, The Journal of Immunology, 198 (2017) 4046-4053.
14.J. Karlberg, D.S. Chong, W.Y. Lai, Do men have a higher case fatality rate of severe acute respiratory syndrome than women do?, Am J Epidemiol, 159 (2004) 229-231.
15.Z. Li, M. Wu, J. Guo, J. Yao, X. Liao, S. Song, M. Han, J. Li, G. Duan, Y. Zhou, X. Wu, Z. Zhou, T. Wang, M. Hu, X. Chen, Y. Fu, C. Lei, H. Dong, Y. Zhou, H. Jia, X. Chen, J. Yan, Caution on Kidney Dysfunctions of 2019-nCoV Patients, medRxiv, (2020).
16.W.H. Ding YQ, Shen H, Li ZG, Geng J, Han HX, Cai JJ, Li X, Kang, W.D. W, Lu YD, Wu DH, He L, Yao KT, The clinical pathology of severe acute respiratory syndrome (SARS): a report from China., J Pathol, 2003, 200:282–289 (2003).
17.Z.L. Lang ZW, Zhang SJ, Meng X, Li JQ, Song CZ, Sun L, Zhou YS, Dwyer DE, A clinicopathological study of three cases of severe acute respiratory syndrome (SARS). , Pathology, 2003, 35:526–531 (2003).
18.C.P. Chong PY, Ling AE, Franks TJ, Tai DY, Leo YS, Kaw GJ,, C.K. Wansaicheong G, Ean Oon LL, Teo ES, Tan KB, Nakajima, S.T. N, Travis WD, Analysis of deaths during the severe acute respiratory syndrome (SARS) epidemic in Singapore: challenges in determining a SARS diagnosis., Arch Pathol Lab Med, 2004,128:195–204 (2004).
19.T.W. Chu KH, Tang CS, Lam MF, Lai FM, To KF, Fung KS, Tang HL, Yan WW, Chan HW, Lai TS, Tong KL, Lai KN, Acute renal impairment in coronavirus-associated severe acute respiratory syndrome., Kidney Int, 2005, 67:698–705 (2005).
20.H.P. Wu VC, Lin WC, Huang JW, Tsai HB, Chen YM, Wu KD, and the SARS Research Group of the National Taiwan, Acute renal failure in SARS patients: more than rhabdomyolysis. , Nephrol Dial Transplant 2004, 19:3180–3182 (2004).




